Step 9 miRNA, lncRNA and circRNA databases
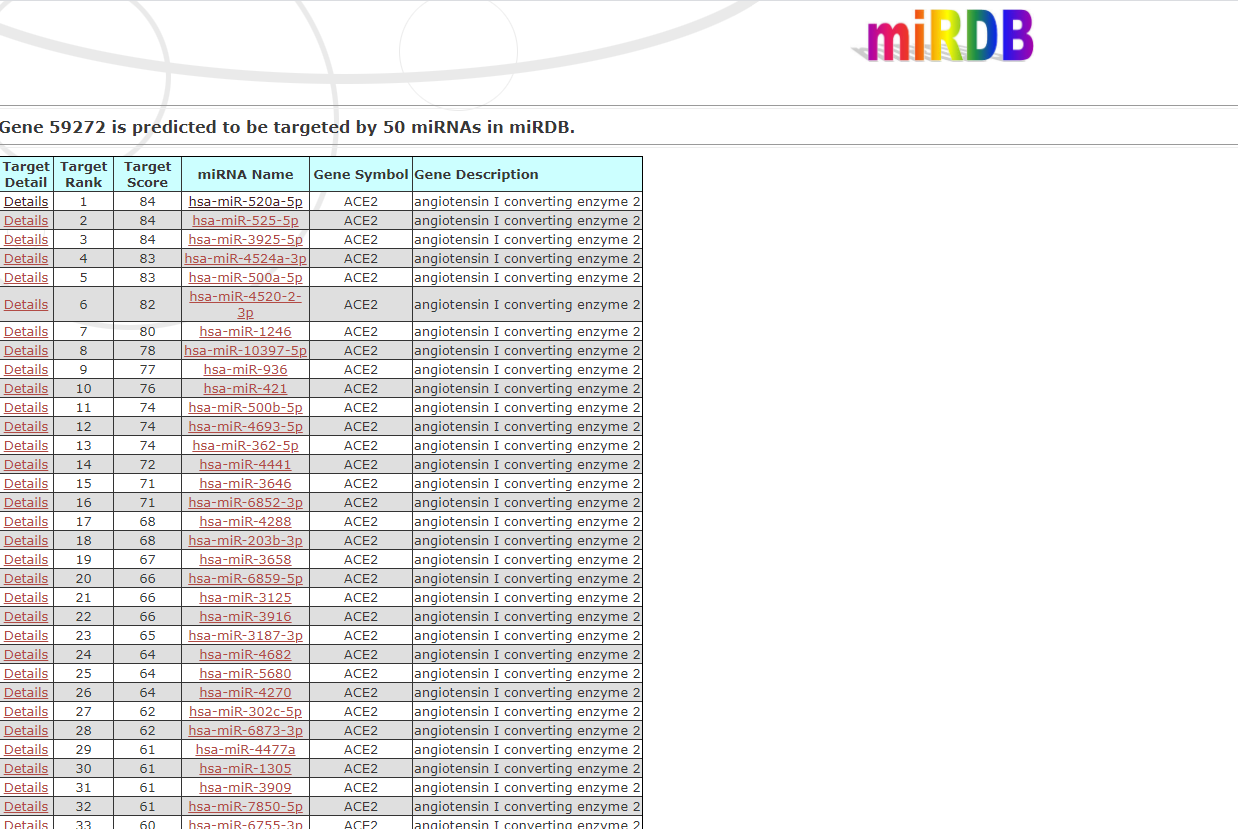
We used one miRNA database, one lncRNA and circRNA database, and one viral miRNA database in order to improve our results. The used miRNA database was miRDB, an online database for miRNA target prediction and functional annotations. All the targets in miRDB were predicted by a bioinformatics tool, MirTarget, which was developed by analyzing thousands of miRNA-target interactions from high-throughput sequencing experiments. miRNAs, as well as associated functional annotations, are presented in the FuncMir Collection in miRDB. The user can choose one of the following search options: Search by miRNA name OR Search by gene target. The initial search returns 50 miRNA candidates to bind the ACE2 gene:
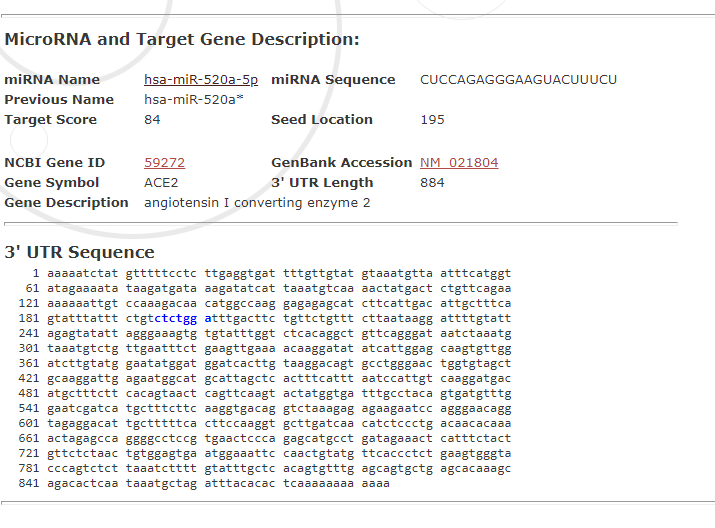
Select the first result, the user can visualize the miRNA target and description:
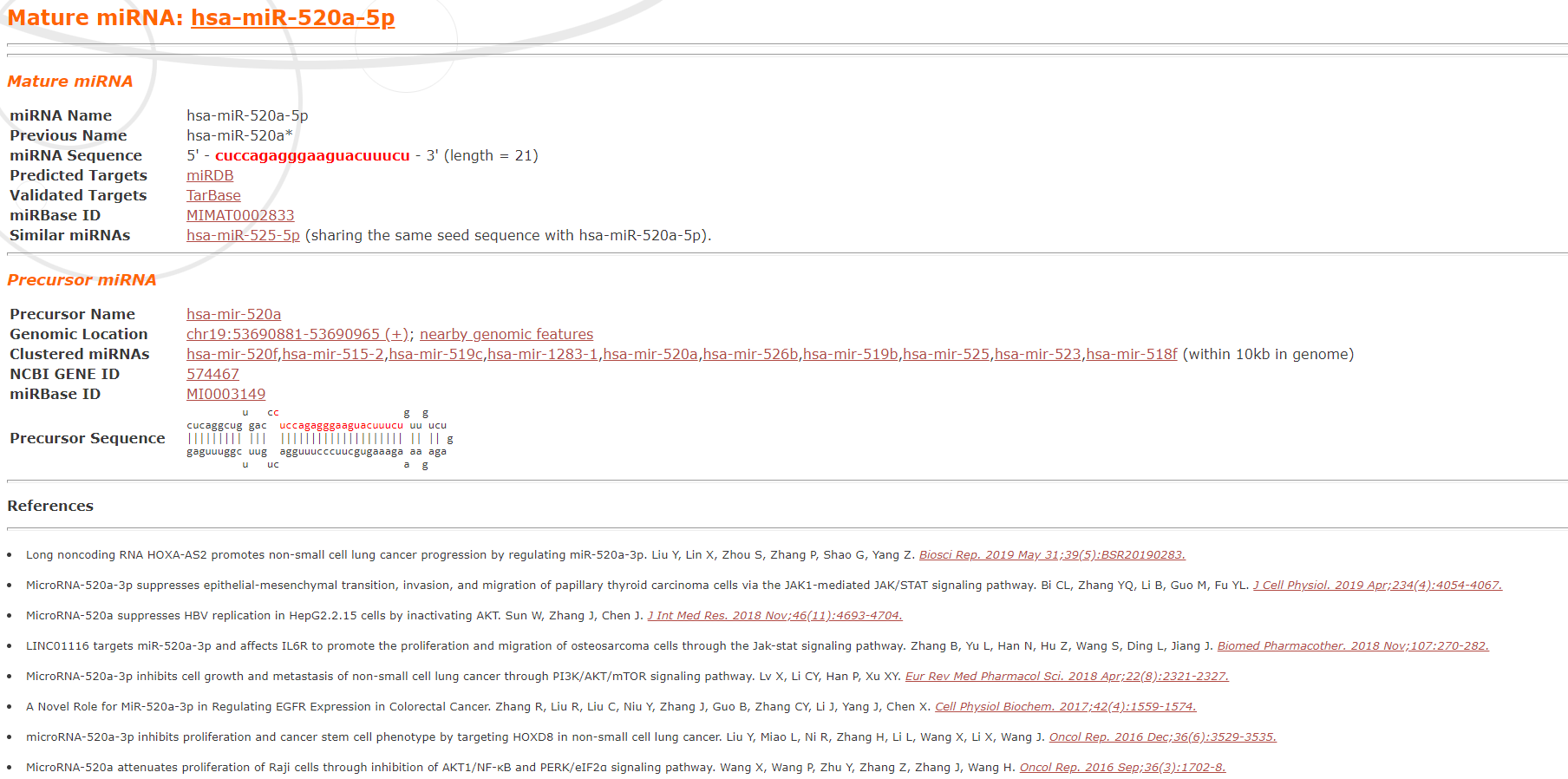
The seed location is marked in blue. The user may access the miRNA information and related references:
For lncRNA and circRNA we used exoRBase, a repository of circular RNA (circRNA), long non-coding RNA (lncRNA), and messenger RNA (mRNA) derived from RNA-seq data analyses of human blood exosomes. Experimental validations from published literature are also included. The database allows the visualization of RNA expression profiles based on normalized RNA-seq data from both normal individuals and patients with different diseases. The user can visualize the data in three different plots: line chart, heatmap, and box plot.
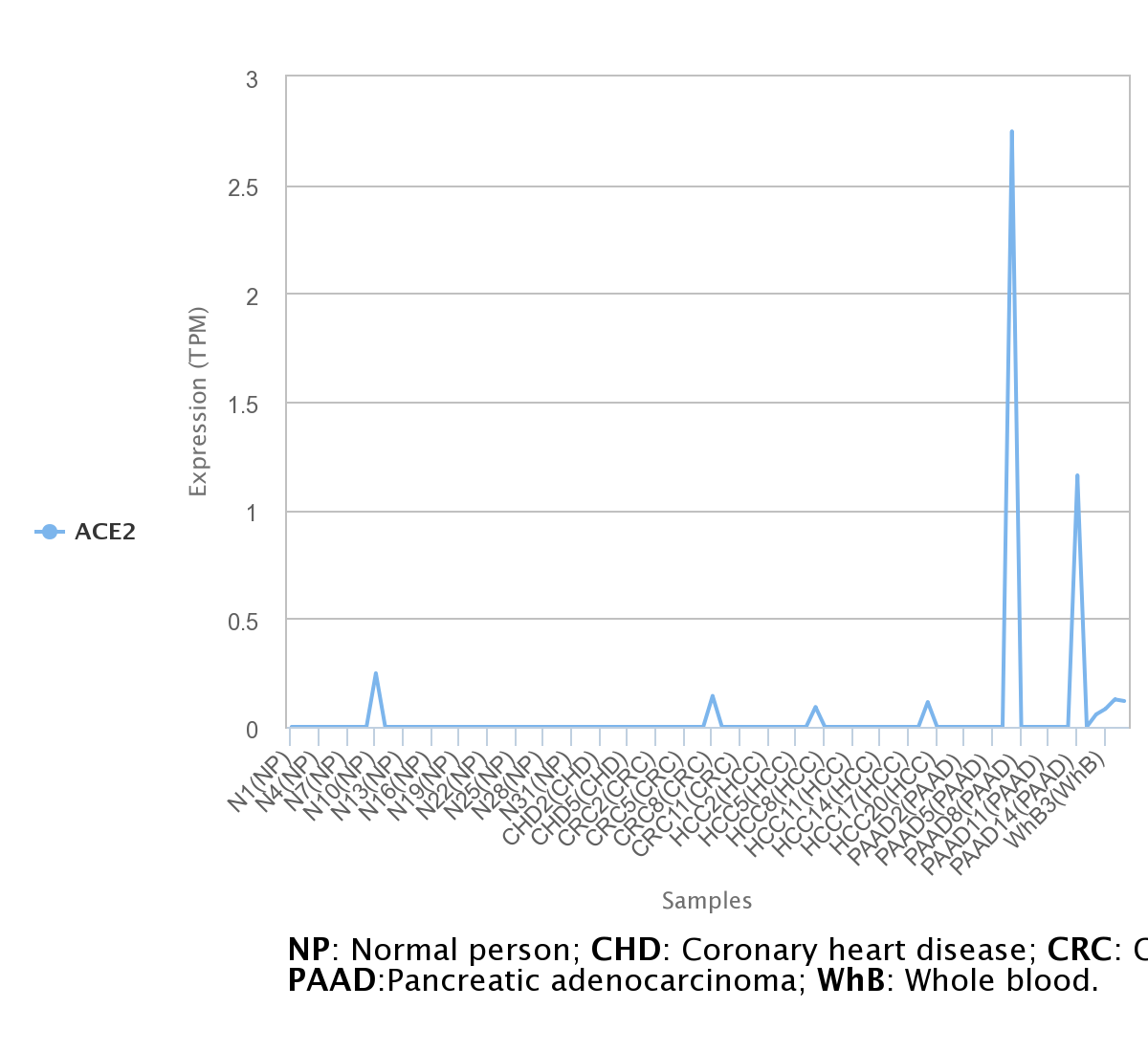
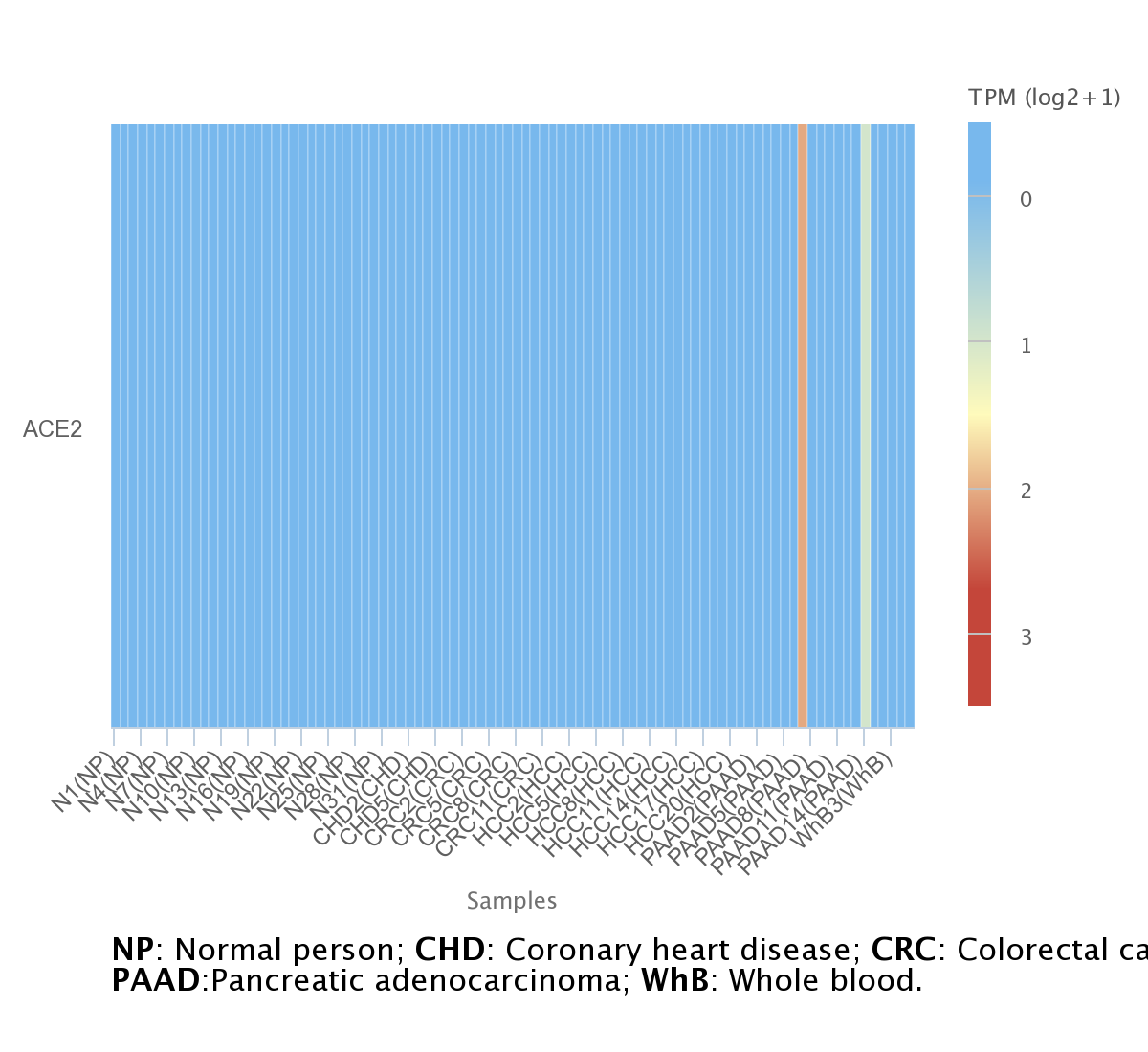
.png)
Here we observe a interesting expression of ACE2 in pancreatic cancer, suggesting that gene is may a important biomarker for the disease.
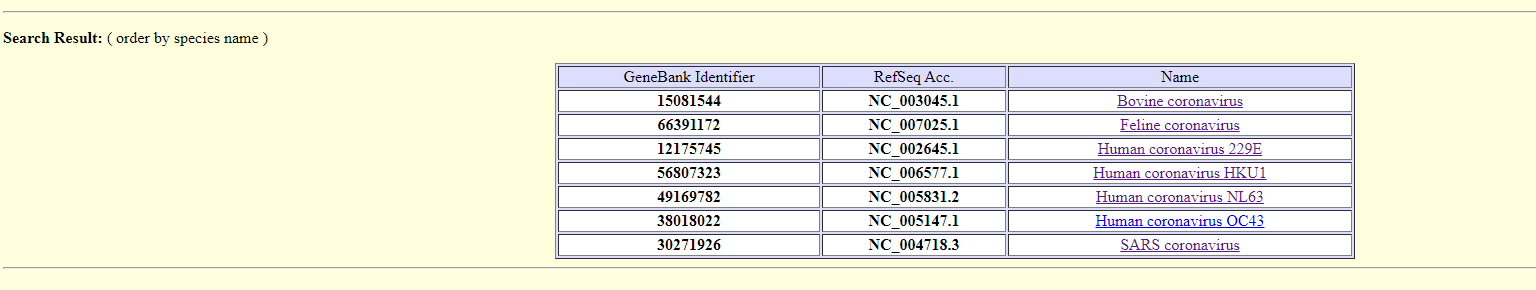
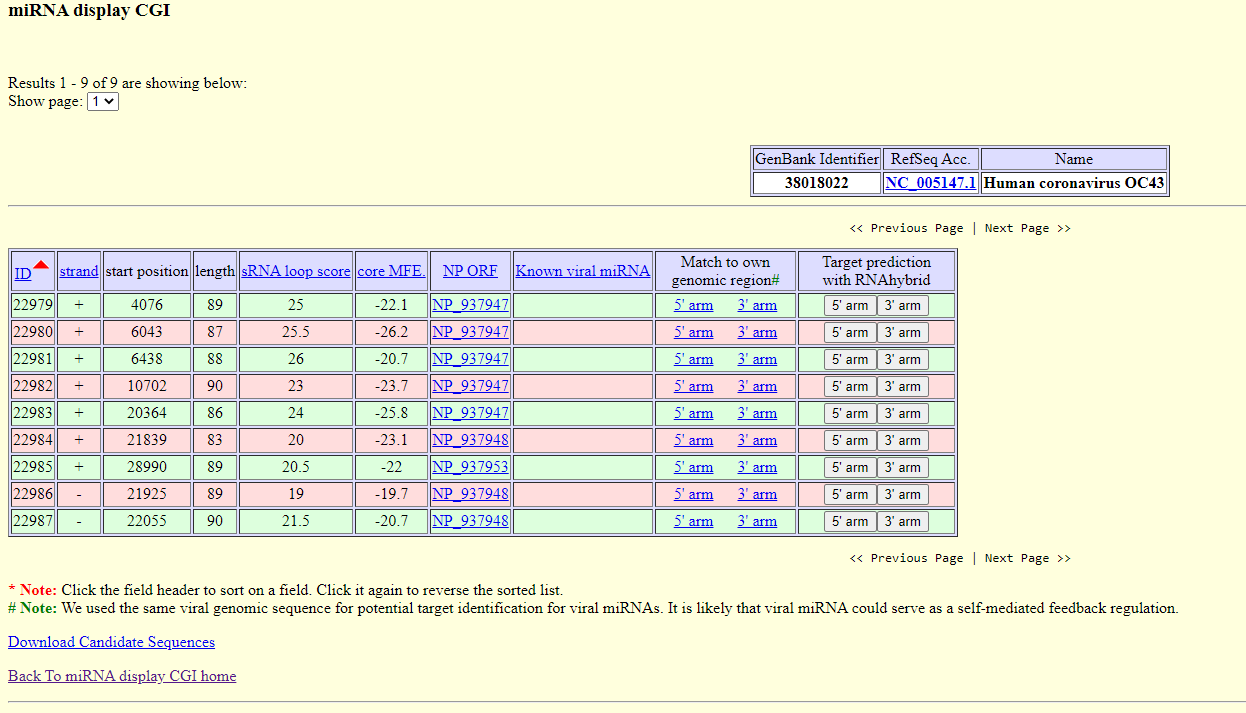
Viral miRNA were analyzed by VirMir, a database with predicted viral miRNA candidate hairpins. The microRNA hairpin discovery pipeline was also applied to discover viral encoded microRNAs. In total, the genomes of 2266 viruses were analyzed. Users may query the putative miRNA hairpins of a specific viral species by the hierarchical menu or by search function using the GenBank Identifier or RefSeq accession number (e.g., 30844336 or NC_003663 for Cowpox virus). In addition, users can also search for the putative target genes of a particular viral miRNA hairpin by an RNAhybrid service link. The 3’-UTR regions of human, mouse, rat, zebrafish, Arabidopsis, and rice genes are available for search. Results:
Exploring Human coronavirus OC43 - first result - 22979
Exploring Human coronavirus OC43 - first result - 22979 Target prediction with RNAhybrid - For each putative or known miRNA, the user may query the target sites in the 3’UTR regions of genes of human, mouse, rat, zebrafish, rice, or Arabidopsis. From the human.rna.fna file downloaded from NCBI, we fetched the sequences of human 3’UTRs based on the CDS (coding sequence) positions acquired from the human.rna.gbff file. We adopted the same strategy for the mouse, rat, zebrafish, rice and Arabidopsis 3’UTRs. In our case, the result sent by e-mail was null, which means there’s no matching sequences returned from RNAhybrid.