Step 10 Metabolic databases
CIDeR is a manually curated database of interactions between disease-related elements such as biomolecules (proteins, metabolites etc.) and other factors (biological processes, phenotypes etc.). The aim of CIDeR is to serve as knowledge base for experimentally-oriented scientists and as resource for bioinformatics applications.
- Per default the search space includes all diseases and all types of data. Queries are not case-sensitive and search all database content that includes the query term (e.g. Mito is sufficient to find everything concerning mitochondria). This type of query is highly unspecific as it includes also information such as gene/protein name synonyms and authors names from referenced literature.
- The first box of the search options (Diseases) allows to restrict the search space for the disease/-s of interest.
- The second box of the search options (Field) allows searching within a specific type of data, e.g. only within genes/proteins, biological processes etc.
- Combined queries can be performed by using Refine query. By clicking on Refine query a second search field is opened. A drop-down menu allows combining the two queries with the Boolean operators AND, OR and NOT.
- The search space can be restricted by setting the search term into double quotes (e.g. “MAPK1” will not find MAPK14 which would happen without quotes)
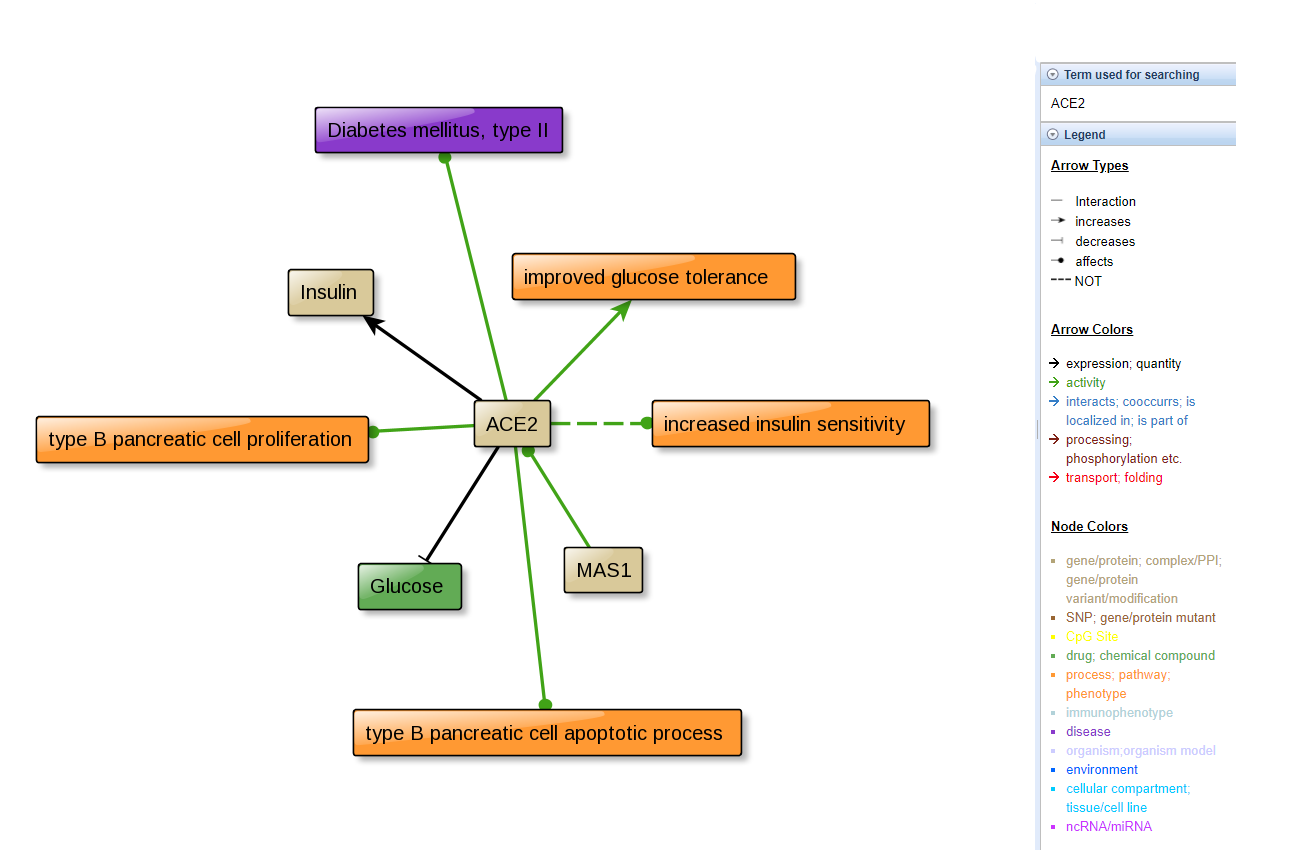
- CIDer have information about several diseases, like Alzheimer disease, Amyotrophic lateral sclerosis, Autism, Bipolar disorder, COXPD, Cancer, Cardiovascular diseases Diabetes mellitus (type II), Epilepsy, Farber lipogranulomatosis, Inflammatory bowel disease, Lung disease, Major depressive disorder, Multiple sclerosis, Muscular atrophy Neutropenia, severe congenital, Other, Parkinson disease, Post-traumatic stress disorder, Postpartum depression, Rare disease, and Schizophrenia. The result of ACE2 input are bellow:
Note: the output only returns diabetes data. The colors are listed in the legend in the right.
On the other hand, HumanMine integrates many types of data for Homo sapiens and Mus musculus. You can run flexible queries, export results and analyze lists of data. This page lists the data that is included in the current release https://www.humanmine.org/humanmine/dataCategories.do. The user can enter names, identifiers, or keywords for genes, proteins, pathways, ontology terms, authors, etc. (e.g., eve, PPARG_HUMAN, glycolysis, ACTN2). The first output for ACE2 was a gene summary, and the user can navigate into several topics: Summary, Function, Genomics, Proteins, SNPs, Disease, Homology, Interactions, Expression, Gene Ontology, Other. However, we got the following error message: “There has been an internal error while processing your request. The problem has been logged and will be investigated. You may also send us an email describing how you encountered this error. The problem may be temporary, in which case you might wish to go back and try your request again, or you might want to go to the home page.”
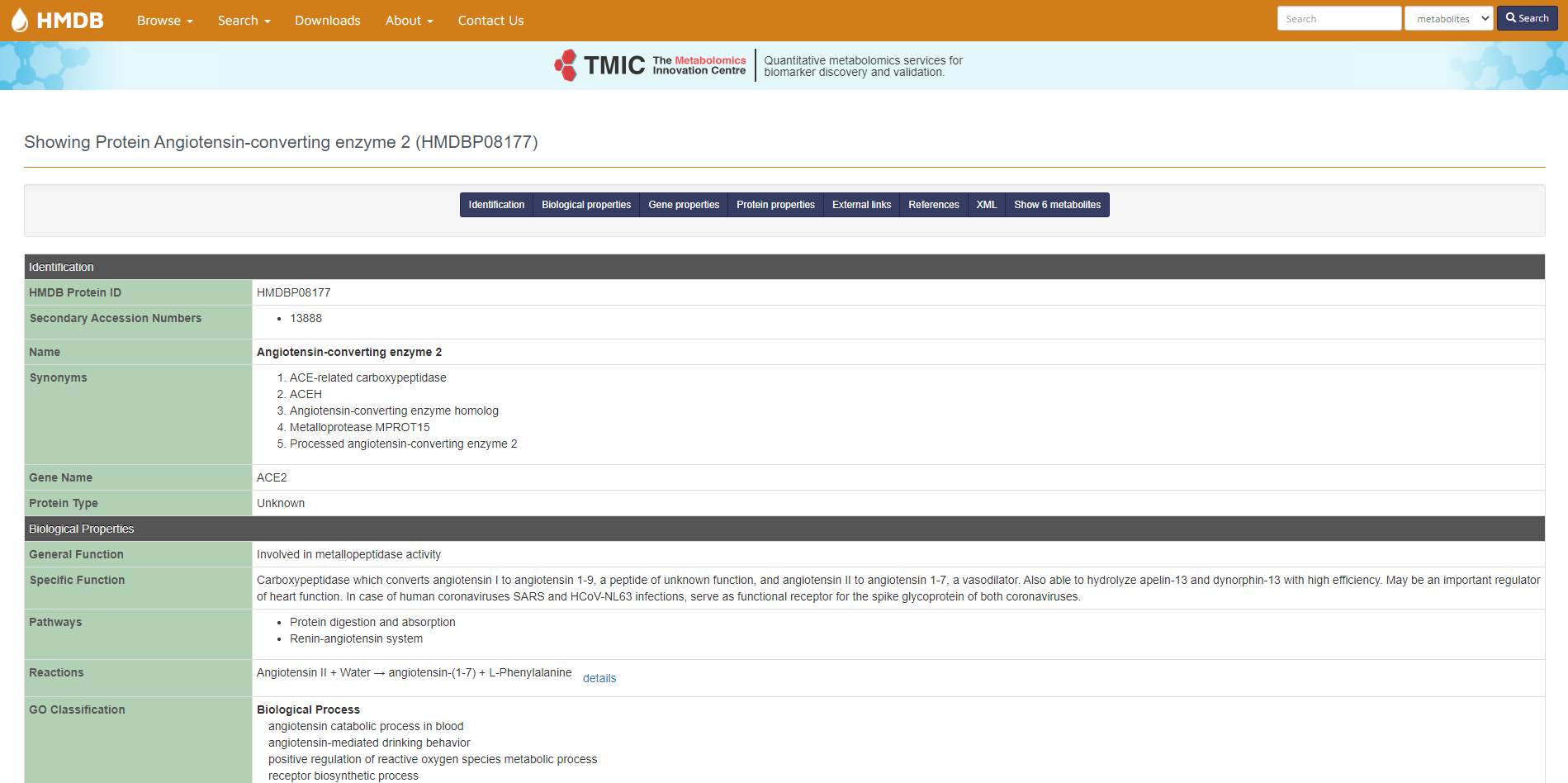
For this reason, we used another metabolic database, The Human Metabolome Database (HMDB). The database contains information about small molecule metabolites found in the human body. It is intended to be used for applications in metabolomics, clinical chemistry, biomarker discovery and general education. The database is designed to contain or link three kinds of data: 1) chemical data, 2) clinical data, and 3) molecular biology/biochemistry data. The database contains 114,221 metabolite entries, including both water-soluble and lipid-soluble metabolites as well as metabolites that would be regarded as either abundant (> 1 uM) or relatively rare (< 1 nM). Additionally, 5,702 protein sequences are linked to these metabolite entries. Each MetaboCard entry contains 130 data fields with 2/3 of the information being devoted to chemical/clinical data and the other 1/3 devoted to enzymatic or biochemical data. Many data fields are hyperlinked to other databases (KEGG, PubChem, MetaCyc, ChEBI, PDB, UniProt, and GenBank) and a variety of structure and pathway viewing applets. The HMDB database supports extensive text, sequence, chemical structure, and relational query searches. The result can also be visualized into SMPDB (The Small Molecule Pathway Database) that is an interactive, visual database containing more than 30 000 small molecule pathways found in humans only.
The result can also visualize into SMPDB (The Small Molecule Pathway Database) is an interactive, visual database containing more than 30 000 small molecule pathways found in humans only.
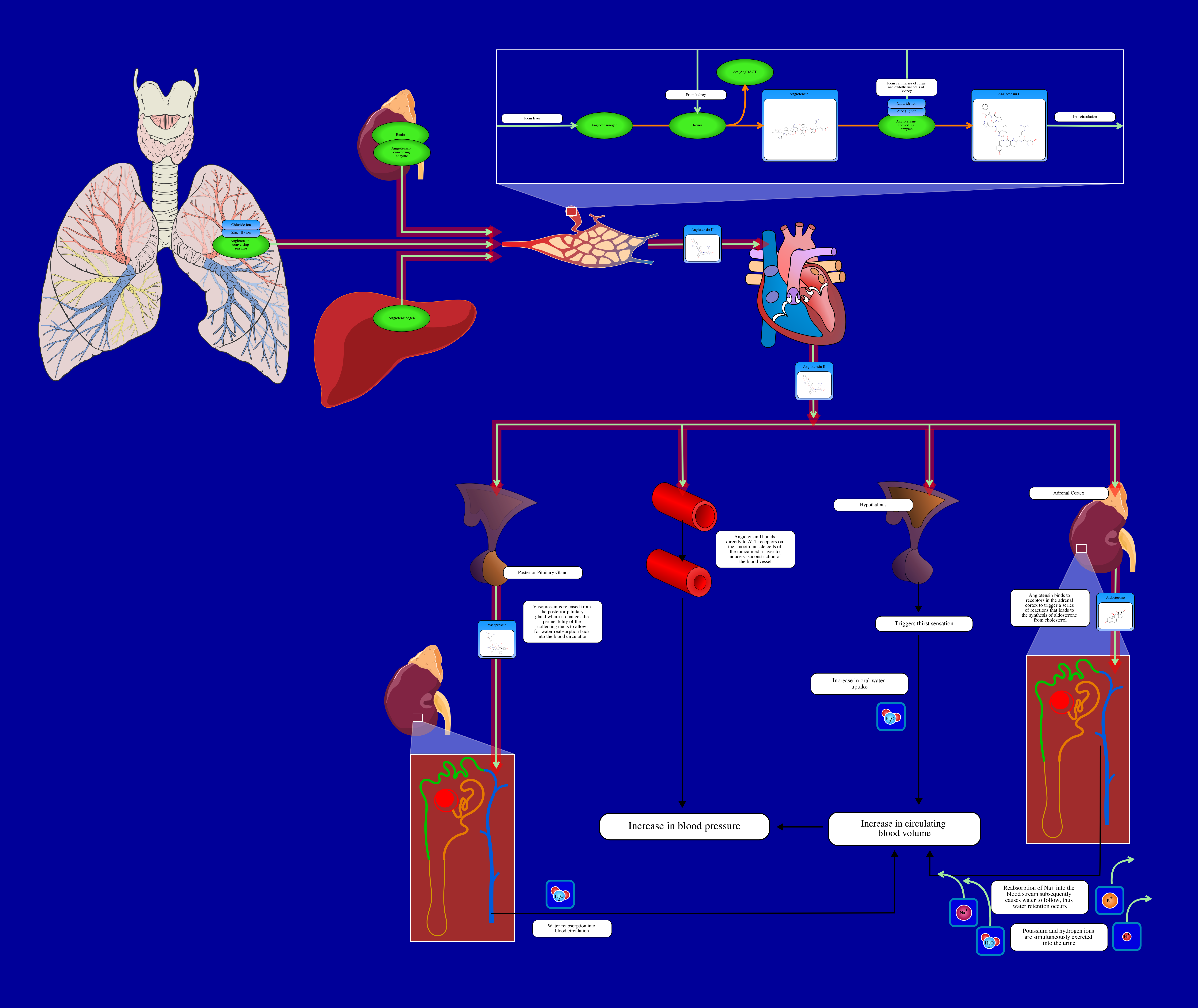
The interactive result was shown in the following adress: https://smpdb.ca/view/SMP0000587