Step 11 Disease-specific and variant-disease association
First we search informations about ACE2 in Varsome - (for more informations, visit the documentation https://docs.varsome.com/introduction).
- Gene basic info
- Region browser
- Structural browser
- Transcripts
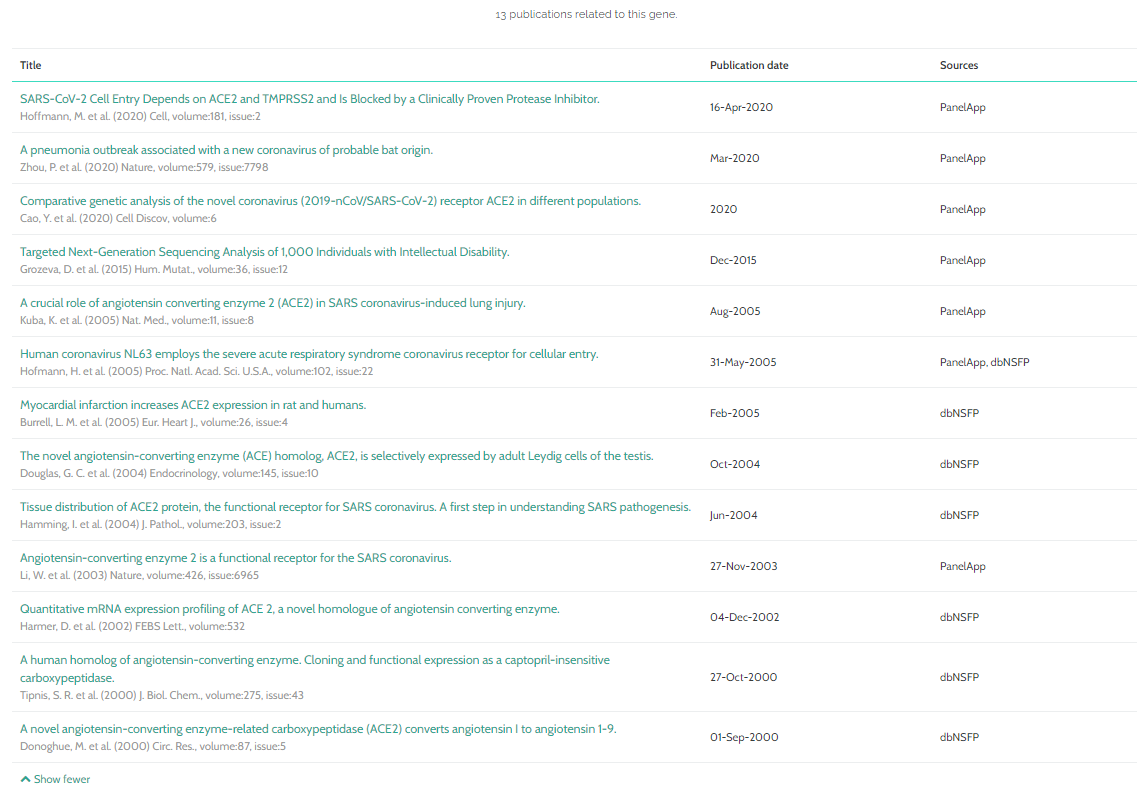
- Publications
- Clinical Statistics
- GTEx
- dbNSFP Genes
- GnomAD Genes
- ExAC genes
- Human Phenotype Ontology
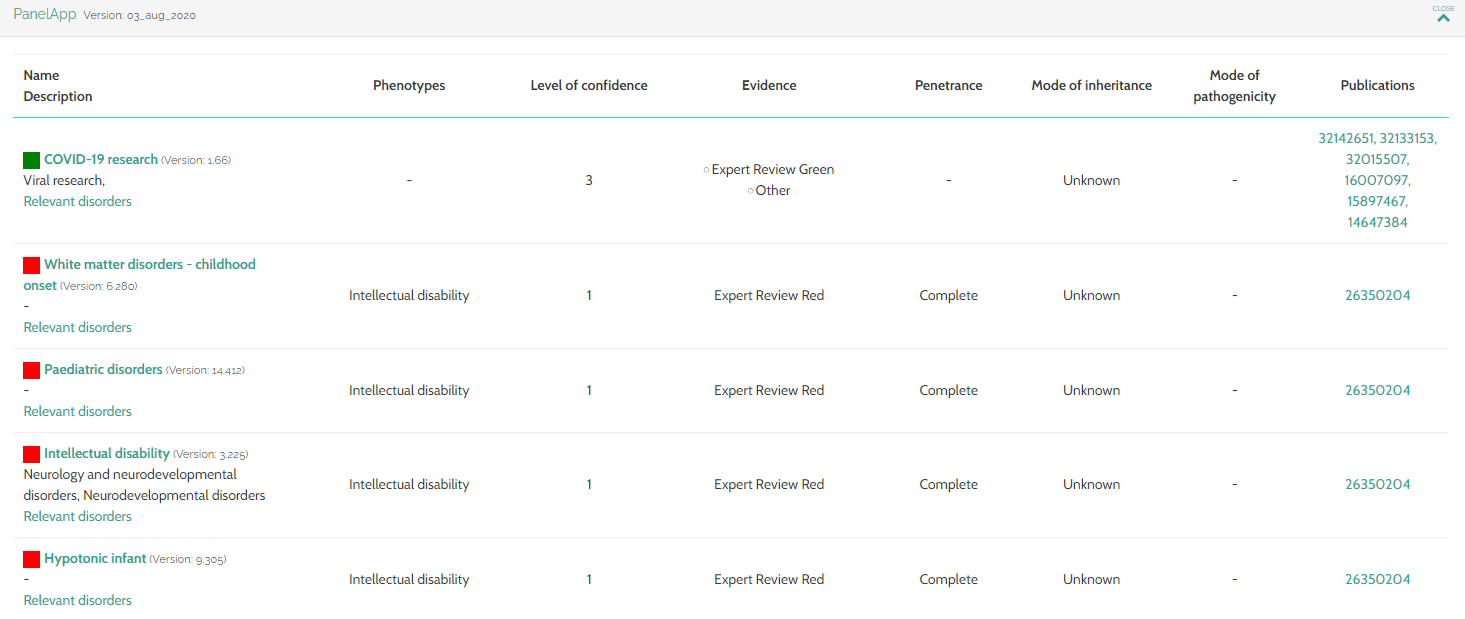
- PanelApp
- PharmGKBVarSome (Premium)
- AACT Clinical TrialsVarSome (Premium)
- DGIVarSome (Premium)
- CPICVarSome (Premium)
Here, we we only show results related to variants:
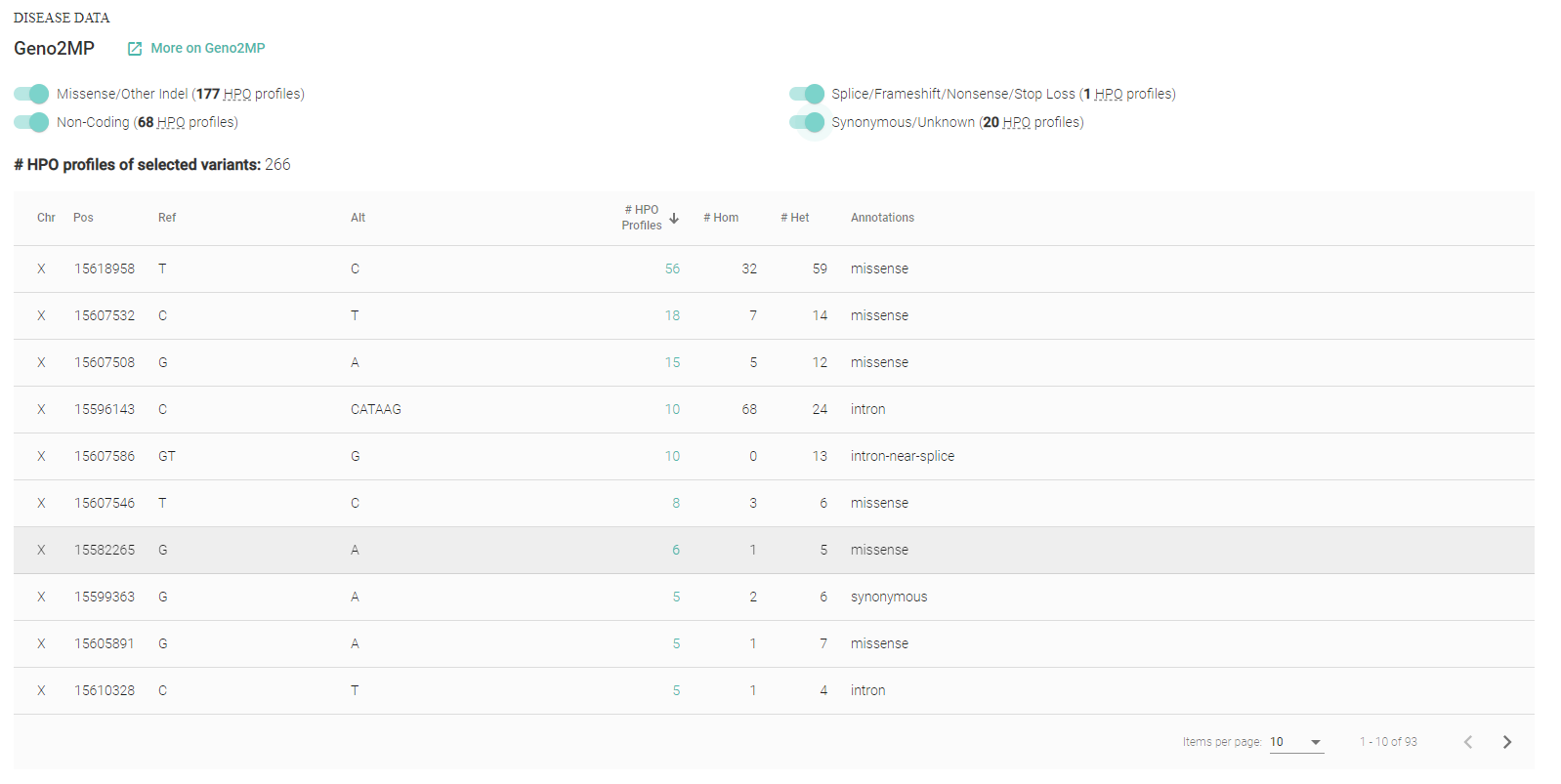
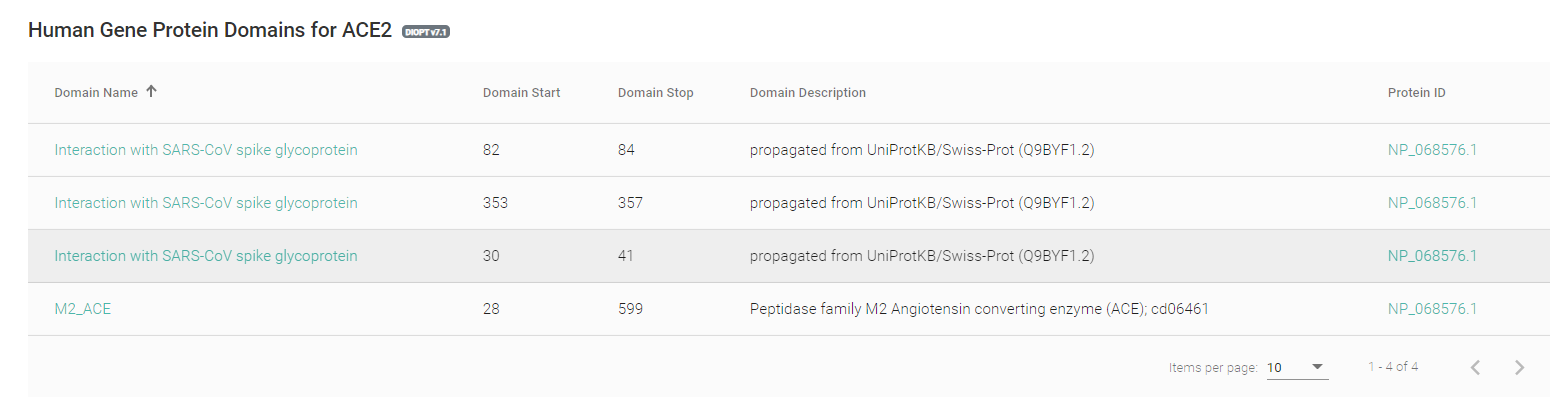
The second database, MARRVEL (Model organism Aggregated Resources for Rare Variant ExpLoration) aims to facilitate the use of public genetic resources to prioritize rare human gene variants for study in model organisms. To facilitate the search process and gather all the data in a simple display we extract data from human data bases (OMIM, ExAC, ClinVar, Geno2MP, DGV, and DECIPHER) for efficient variant prioritization. The protein sequences for eight organisms (S. cerevisiae, S. pombe, C. elegans, D. melanogaster, D. rerio, M. musculus, R. norvegicus, and H. sapiens) are aligned with highlighted protein domain information via collaboration with DIOPT. The key biological and genetic features are then extracted from existing model organism databases (SGD, PomBase, WormBase, FlyBase, ZFIN, MGI, and RGD).
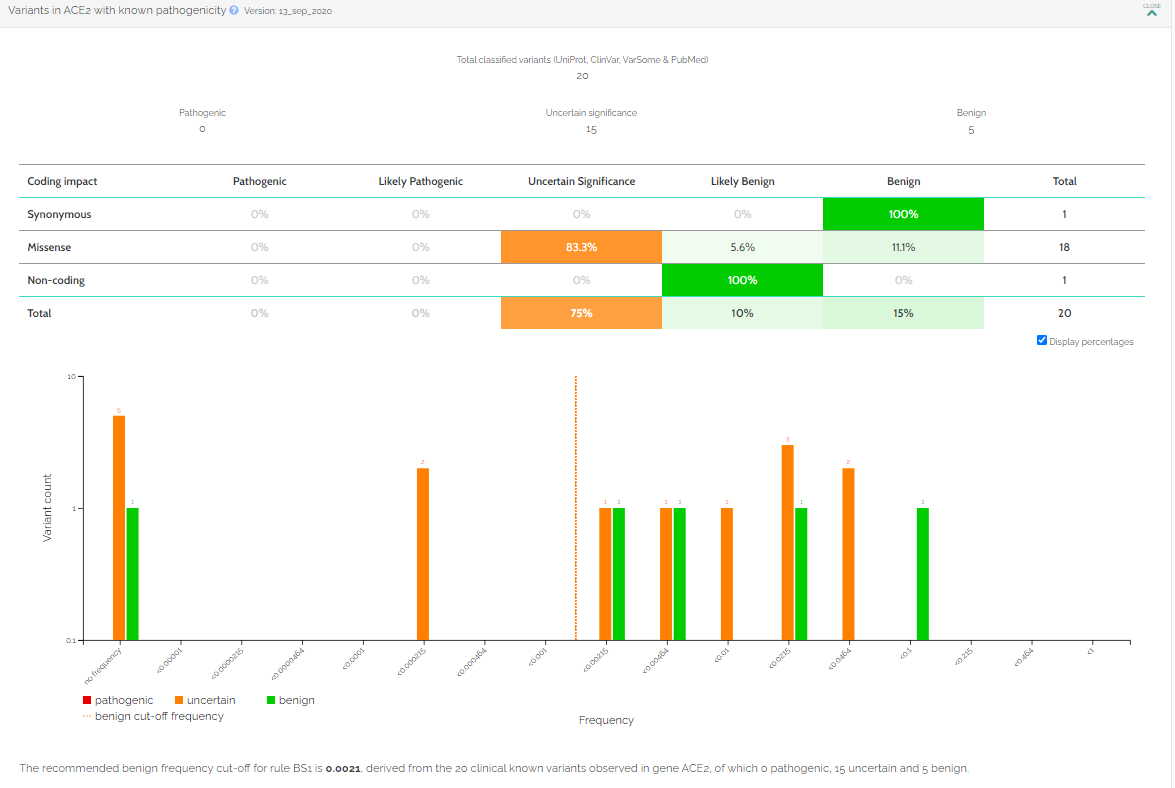
Disease / variant data

Additionally, the database provides information about expression of Ortholog Genes of ACE2 and your gene ontology terms:
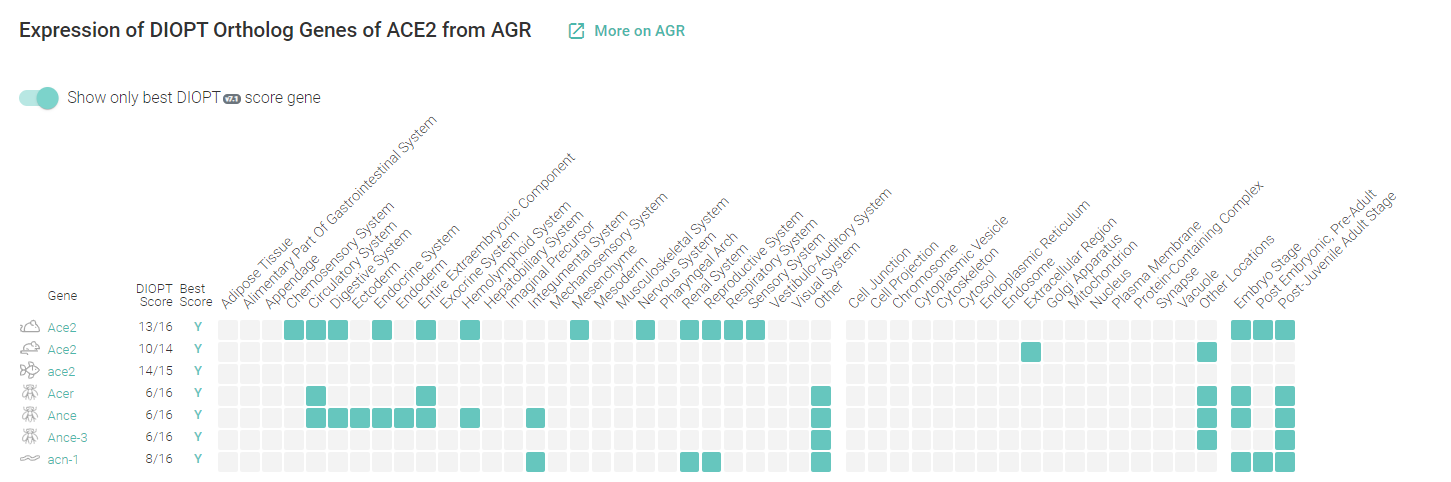
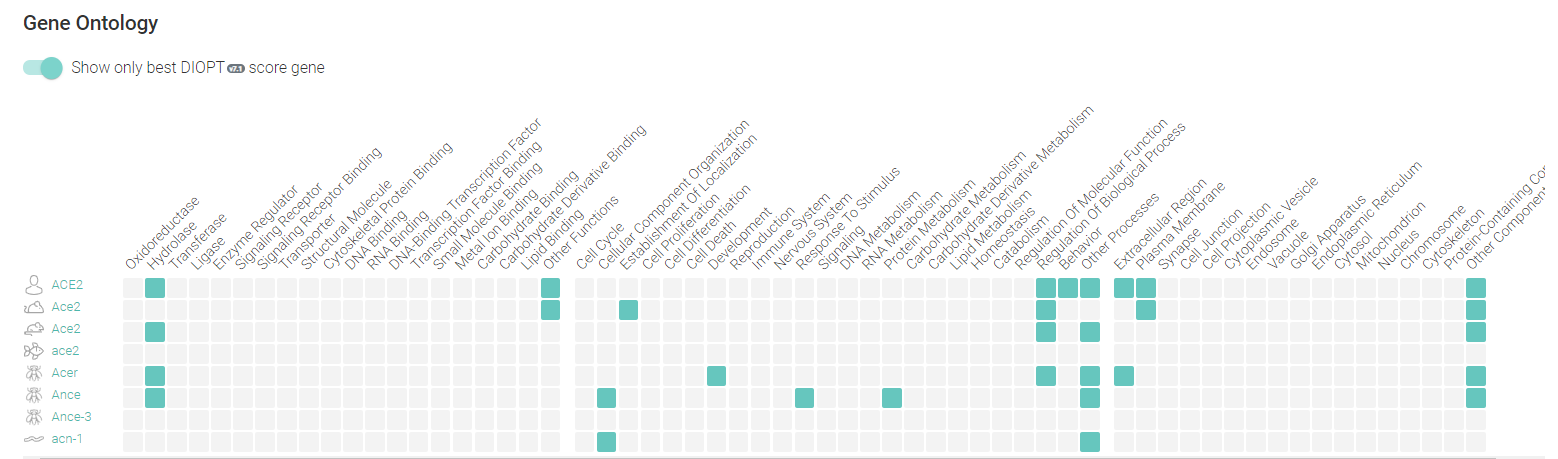
The two used variant-disease databases are complementary and user-friendly. With the databases presented here, the user can explore the human ACE2 variations across different populations and studies.