Step 5 Comparative databases
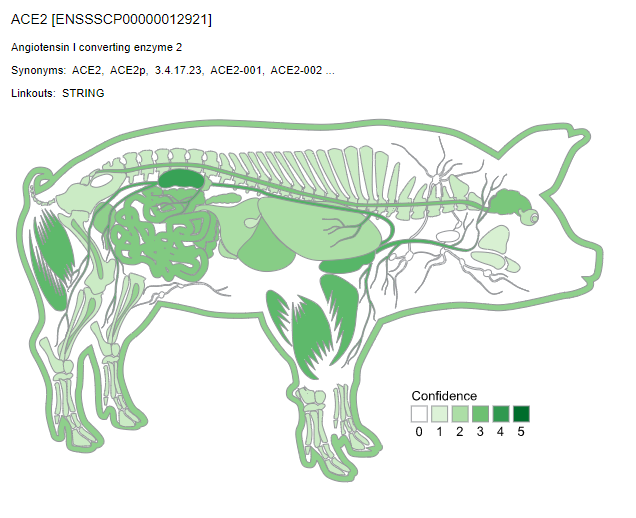
We used TISSUES database to compare the ACE2 gene expression across human and animal models data.
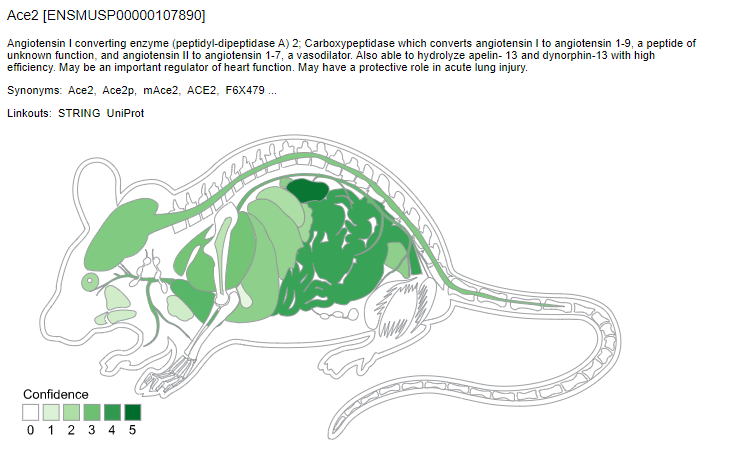
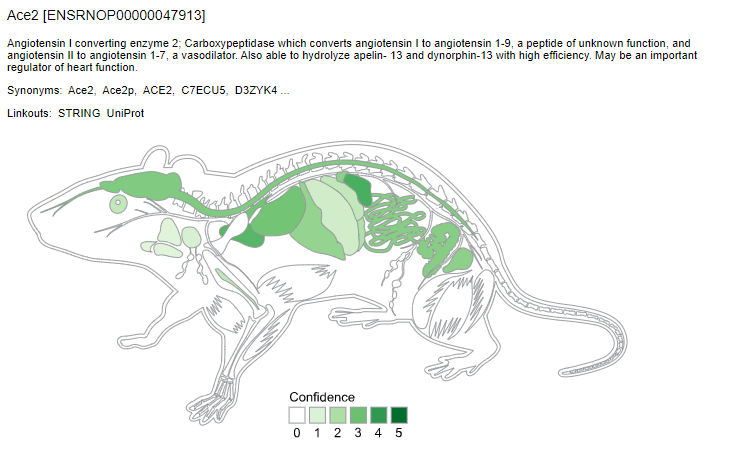
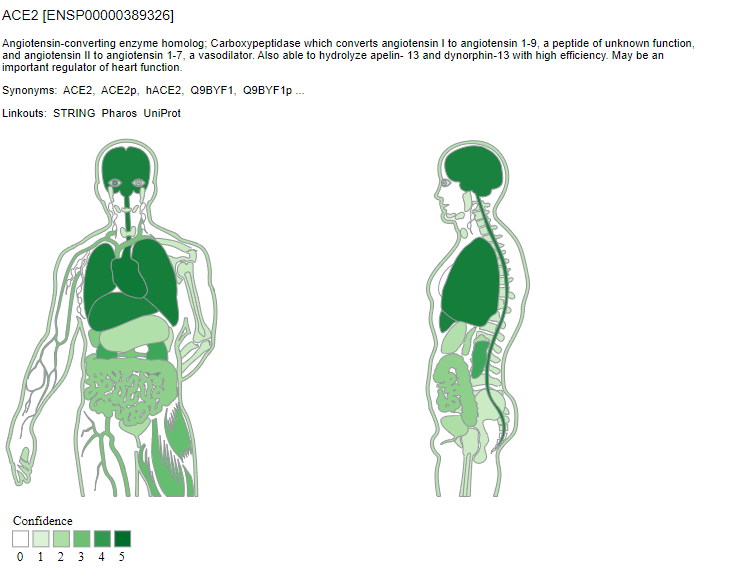
TISSUES also provides experimental curated data, text mining interactions, and orthologs and paralogs. The confidence of each association is given by stars, where ★★★★★ is the highest confidence and ★☆☆☆☆ is the lowest. We can observe the differential expression pattern of ACE2 for each animal model. This may suggest that the choice of animal models to study ACE2 expression levels would have to take these parameters into account.
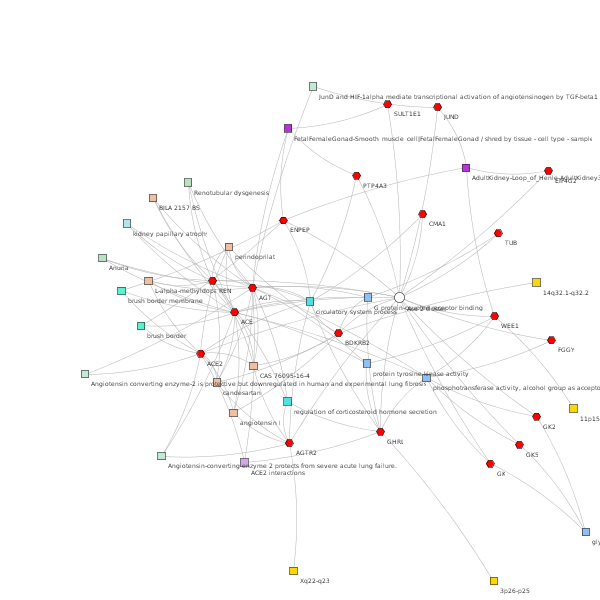
For the next step, we search for a list of genes that interact with ACE2 using the GeneMania web https://genemania.org/. The search returns the following gene list: ACE2, GK, ACE, AGT, CTD-2501B8.1, TMEM27, AGTR2, REN, SULT1E1, RP11-668G10.2, GK2, BDKRB2, EIF4G2, ENPEP, PTP4A3, WEE1, GK5, GHRL, JUND, TUB, SERPIND1, FGGY, CMA1, AQP8 . We use this gene list as input into TopCluster web server https://toppcluster.cchmc.org/. The next step shows a feature to explore the data (e.g., gene ontology, drugs, Pubmed, related diseases) and statistical options, such as correction methods (we choose Bonferroni) and p-value cutoff (0.05). Besides, we may choose to start the analysis in the Functional Enrichment module or Interaction Network. The database also provides the genes found in the gene list. In our case, 3 genes were not found in the database. We select the top score interactions to run the next step of the analysis.