Step 2 Searching in genomic and sequence databases
To obtain more informations about ACE2 related diseases, we search informations in DisGenet and Harmonizome. The initial screen of DisgeNet shows three options of input: 1) disease 2) genes 3) variants We choose the second option: genes to search information about the ACE2 gene. The screen shows these options:
- Entrez Identifier: 59272
- Gene Symbol: ACE2
- Uniprot Accession: Q9BYF1
- Full Name: Angiotensin I converting enzyme 2
- Protein Class: Enzyme
- DPI: 0.769
- DSI: 0.477
- pLi: 0.99769
Where DPI is the Disease Pleotropy index, the DPI ranges from 0 to 1. Example: gene KCNT1 is associated to 39 diseases, 4 disease groups, and 18 phenotypes. 29 out of the 39 diseases have a MeSH disease class. The 29 diseases are associated to 5 different MeSH classes. The DPI index for KCNT1 = 5/29 ~ 0.172. Nevertheless, gene APOE, associated to more than 700 diseases of 27 disease classes has a DPI of 0.931. If the gene/variant has no DPI value, it implies that the gene/variant is associated only to phenotypes, or that the associated diseases do not map to any MeSH classes.
DSI is the Disease Specificity Index, which consists into There are genes (or variants) that are associated wiht multiple diseases (e.g. TNF) while others are associated with a small set of diseases or even to a single disease. The Disease Specificity Index (DSI) is a measure of this property of the genes (and variants). It reflects if a gene (or variant) is associated to several or fewer diseases. he DSI ranges from 0.25 to 1. Example: TNF, associated to more than 1,500 diseases, has a DSI of 0.263, while HCN2 is associated to one disease, with a DSI of 1. If the DSI is empty, it implies that the gene/variant is associated only to phenotypes.
And pLi is a GNOMAD https://gnomad.broadinstitute.org/ metric is the probability of being loss-of-function intolerant, is a gene constraint metric provided by the GNOMAD consortium. A gene constraint metric aims at measuring how the naturally occurring LoF (loss of function) variation has been depleted from a gene by natural selection (in other words, how intolerant is a gene to LoF variation). LoF intolerant genes will have a high pLI value (>=0.9), while LoF tolerant genes will have low pLI values (<=0.1). The LoF variants considered are nonsense and essential splice site variants. In the ACE2 case, the pLi value was 0.99769, indicating that the gene was a loss-of-function intolerant.
The ouput screen also show more options, such as Summary of Gene-Disease Associations, Evidence for Gene-Disease Associations, Summary of Variant-Disease Associations and Evidences of Variant-Disease Associations. Currently, the database also provides a COVID-19 related search https://www.disgenet.org/covid/genes/evidences/?gene_id=59272
In Harmonizome, the ouput of the search for ACE2 gene generates a list of associated terms, such as Name, Description, Synonyms, Proteins, NCBI Gene ID, an option to Download the Associations in a .csv file, Predicted Functions, Co-expressed Genes , and Expression in Tissues and Cell Lines. Image of the plot of Tissue Expression option:
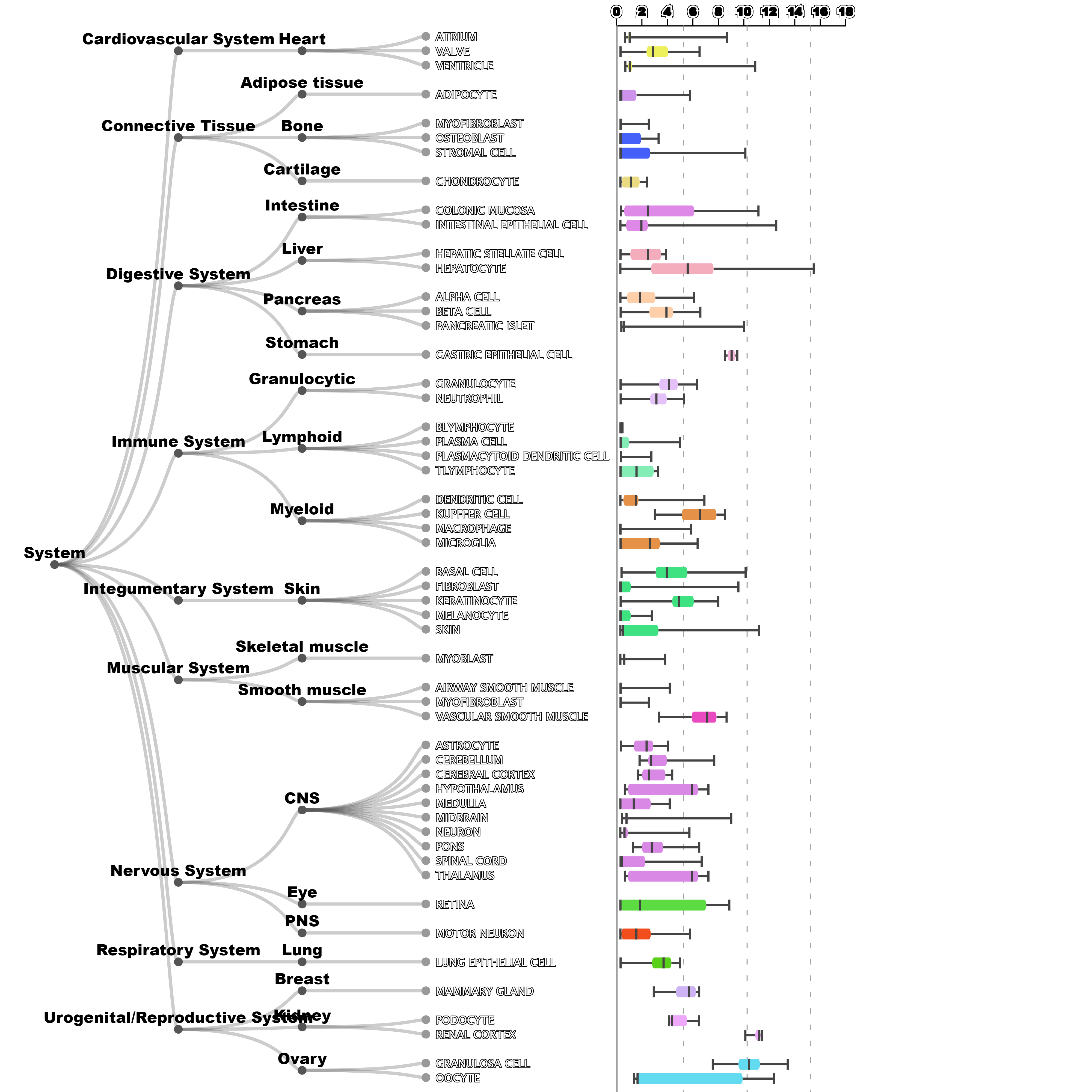
The organ- and cell-specific expression of ACE2 gene suggests that it may play a role in the regulation of cardiovascular and renal function, as well as fertility. In addition, the encoded protein is a functional receptor for the spike glycoprotein of the human coronaviruses SARS and HCoV-NL63.
For Functional Associations, Harmonizome links up to 30 different resources related to the query gene.